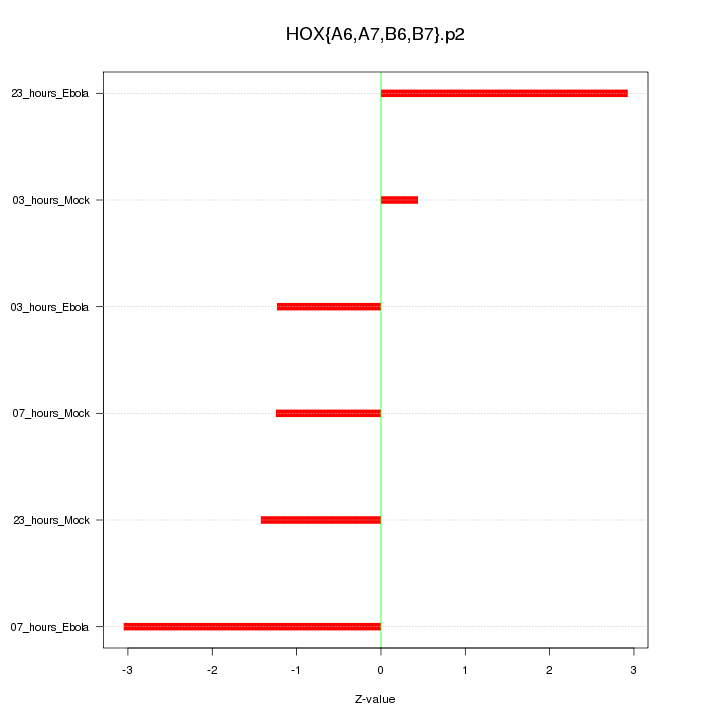
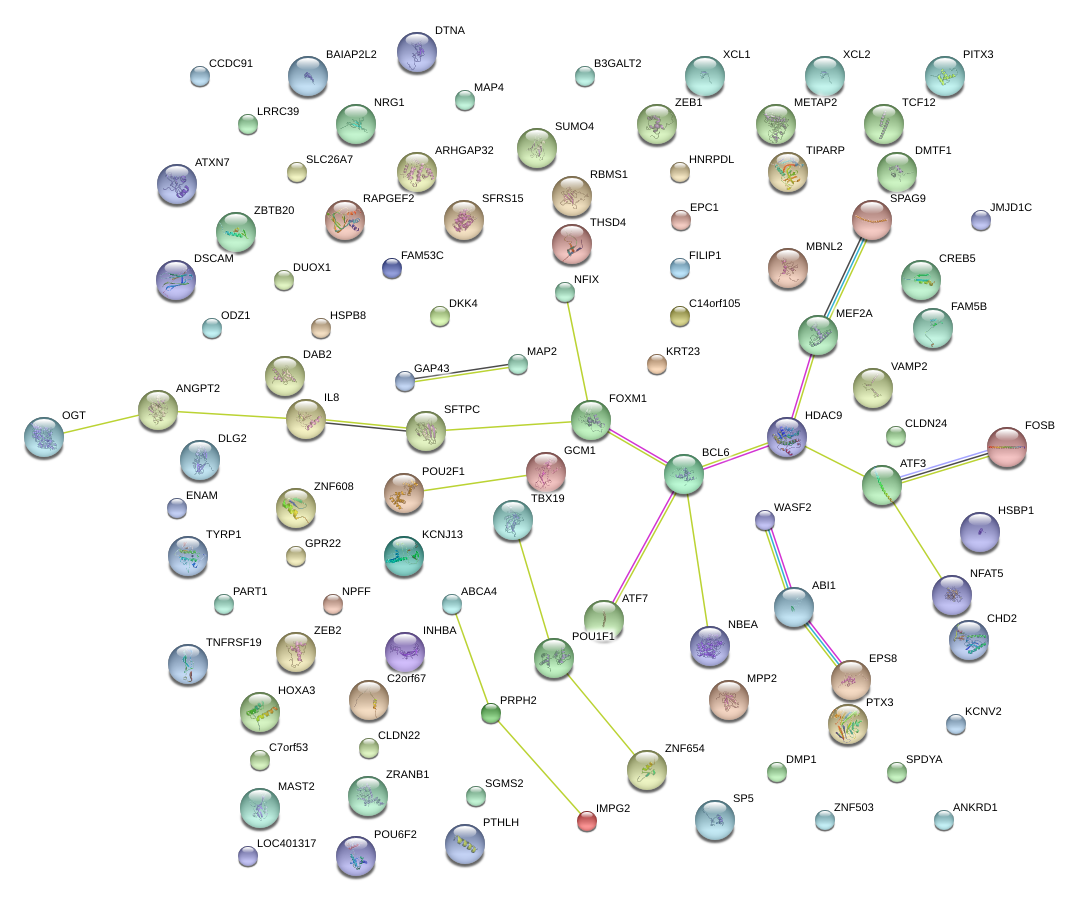
|
chr7_+_18535351
|
7.896
|
NM_001204145
NM_001204146
NM_014707
NM_178423
|
HDAC9
|
histone deacetylase 9
|
|
chrM_+_10123
|
6.952
|
|
|
|
|
chr7_+_28452143
|
6.146
|
NM_182898
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr10_-_92681025
|
4.108
|
NM_014391
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle)
|
|
chr4_-_184243578
|
3.915
|
NM_001185149
|
CLDN24
|
claudin 24
|
|
chr3_-_114478117
|
3.896
|
NM_001164344
NM_001164345
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr15_+_71839540
|
3.786
|
|
THSD4
|
thrombospondin, type I, domain containing 4
|
|
chr1_+_168250277
|
3.707
|
NM_005149
|
TBX19
|
T-box 19
|
|
chr4_+_71494460
|
3.675
|
NM_031889
|
ENAM
|
enamelin
|
|
chr1_+_167298382
|
3.602
|
|
POU2F1
|
POU class 2 homeobox 1
|
|
chrX_-_124097601
|
3.291
|
NM_001163278
NM_001163279
NM_014253
|
ODZ1
|
odz, odd Oz/ten-m homolog 1 (Drosophila)
|
|
chr2_-_145277568
|
3.261
|
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr7_+_28475233
|
3.160
|
NM_004904
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr1_+_167298280
|
3.157
|
NM_001198783
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr8_-_42234562
|
3.129
|
NM_014420
|
DKK4
|
dickkopf homolog 4 (Xenopus laevis)
|
|
chr13_+_36050885
|
3.100
|
NM_001204197
|
NBEA
|
neurobeachin
|
|
chr7_+_18330044
|
3.081
|
|
HDAC9
|
histone deacetylase 9
|
|
chr2_-_145277879
|
3.045
|
NM_001171653
NM_014795
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr4_+_108814419
|
3.019
|
NM_001136257
NM_152621
|
SGMS2
|
sphingomyelin synthase 2
|
|
chr10_-_32667543
|
2.902
|
|
EPC1
|
enhancer of polycomb homolog 1 (Drosophila)
|
|
chr17_-_39092714
|
2.885
|
|
KRT23
|
keratin 23 (histone deacetylase inducible)
|
|
chr1_+_168545710
|
2.775
|
NM_002995
|
XCL1
|
chemokine (C motif) ligand 1
|
|
chr6_-_76203256
|
2.773
|
NM_015687
|
FILIP1
|
filamin A interacting protein 1
|
|
chr3_-_114790221
|
2.770
|
NM_001164343
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr7_+_18535817
|
2.701
|
NM_058176
NM_178425
|
HDAC9
|
histone deacetylase 9
|
|
chr16_+_69680959
|
2.581
|
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr15_+_93443559
|
2.480
|
|
CHD2
|
chromodomain helicase DNA binding protein 2
|
|
chr15_+_93443418
|
2.433
|
NM_001042572
NM_001271
|
CHD2
|
chromodomain helicase DNA binding protein 2
|
|
chr10_-_92680784
|
2.429
|
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle)
|
|
chr17_-_39093671
|
2.356
|
NM_015515
|
KRT23
|
keratin 23 (histone deacetylase inducible)
|
|
chr9_+_12693385
|
2.296
|
NM_000550
|
TYRP1
|
tyrosinase-related protein 1
|
|
chr6_+_149721494
|
2.294
|
NM_001002255
|
SUMO4
|
SMT3 suppressor of mif two 3 homolog 4 (S. cerevisiae)
|
|
chr2_+_210444364
|
2.277
|
NM_002374
NM_031845
NM_031847
|
MAP2
|
microtubule-associated protein 2
|
|
chr7_+_107110501
|
2.254
|
NM_005295
|
GPR22
|
G protein-coupled receptor 22
|
|
chr1_+_177140062
|
2.236
|
|
FAM5B
|
family with sequence similarity 5, member B
|
|
chr3_-_101039403
|
2.236
|
NM_016247
|
IMPG2
|
interphotoreceptor matrix proteoglycan 2
|
|
chr8_-_6420454
|
2.206
|
NM_001118887
NM_001118888
NM_001147
|
ANGPT2
|
angiopoietin 2
|
|
chr4_-_83347778
|
2.187
|
|
HNRPDL
|
heterogeneous nuclear ribonucleoprotein D-like
|
|
chr19_+_13134782
|
2.182
|
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr11_-_129062092
|
2.117
|
NM_001142685
|
ARHGAP32
|
Rho GTPase activating protein 32
|
|
chr6_-_53013581
|
2.074
|
NM_003643
|
GCM1
|
glial cells missing homolog 1 (Drosophila)
|
|
chr2_-_233641225
|
2.007
|
NM_001172416
NM_001172417
NM_002242
|
KCNJ13
|
potassium inwardly-rectifying channel, subfamily J, member 13
|
|
chr10_+_126630691
|
1.979
|
NM_017580
|
ZRANB1
|
zinc finger, RAN-binding domain containing 1
|
|
chr2_-_161130151
|
1.964
|
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1
|
|
chr12_-_53901371
|
1.952
|
NM_003717
|
NPFF
|
neuropeptide FF-amide peptide precursor
|
|
chr2_+_171571800
|
1.925
|
NM_001003845
|
SP5
|
Sp5 transcription factor
|
|
chr3_+_88188261
|
1.911
|
NM_018293
|
ZNF654
|
zinc finger protein 654
|
|
chr1_+_212782103
|
1.855
|
|
ATF3
|
activating transcription factor 3
|
|
chr1_-_193155723
|
1.840
|
NM_003783
|
B3GALT2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2
|
|
chr5_+_137673223
|
1.823
|
NM_001135647
|
FAM53C
|
family with sequence similarity 53, member C
|
|
chr7_-_27166638
|
1.817
|
NM_153631
|
HOXA3
|
homeobox A3
|
|
chr15_+_100173182
|
1.815
|
NM_001130928
|
MEF2A
|
myocyte enhancer factor 2A
|
|
chr14_-_57960432
|
1.815
|
NM_018168
|
C14orf105
|
chromosome 14 open reading frame 105
|
|
chr3_-_187452669
|
1.812
|
NM_001134738
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr1_+_212781969
|
1.789
|
NM_001040619
NM_001206484
NM_001206488
NM_001674
|
ATF3
|
activating transcription factor 3
|
|
chr12_+_119616594
|
1.758
|
NM_014365
|
HSPB8
|
heat shock 22kDa protein 8
|
|
chr12_+_28410132
|
1.755
|
NM_018318
|
CCDC91
|
coiled-coil domain containing 91
|
|
chr3_-_114343052
|
1.739
|
NM_001164347
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr2_+_29038862
|
1.734
|
NM_001008779
|
SPDYA
|
speedy homolog A (Xenopus laevis)
|
|
chr1_-_27732069
|
1.722
|
|
WASF2
|
WAS protein family, member 2
|
|
chr3_+_63953419
|
1.719
|
NM_001128149
|
ATXN7
|
ataxin 7
|
|
chr9_-_5968367
|
1.714
|
|
|
|
|
chr13_+_97874704
|
1.674
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr7_+_112120907
|
1.657
|
NM_001134468
NM_182597
|
C7orf53
|
chromosome 7 open reading frame 53
|
|
chr7_+_28725597
|
1.605
|
NM_001011666
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr5_+_59784004
|
1.599
|
|
PART1
|
prostate androgen-regulated transcript 1 (non-protein coding)
|
|
chr7_+_28448970
|
1.596
|
|
LOC401317
|
uncharacterized LOC401317
|
|
chr18_+_32173281
|
1.588
|
NM_001128175
NM_001198938
NM_001198939
NM_001198940
|
DTNA
|
dystrobrevin, alpha
|
|
chr12_-_28124915
|
1.579
|
NM_002820
NM_198965
|
PTHLH
|
parathyroid hormone-like hormone
|
|
chr18_+_32290195
|
1.575
|
NM_001198941
NM_032978
|
DTNA
|
dystrobrevin, alpha
|
|
chr12_-_15865847
|
1.565
|
|
EPS8
|
epidermal growth factor receptor pathway substrate 8
|
|
chr10_-_65028825
|
1.563
|
NM_004241
|
JMJD1C
|
jumonji domain containing 1C
|
|
chr2_+_210444154
|
1.563
|
|
MAP2
|
microtubule-associated protein 2
|
|
chr12_-_53994783
|
1.535
|
NM_001130059
|
ATF7
|
activating transcription factor 7
|
|
chr19_+_45971249
|
1.515
|
NM_001114171
NM_006732
|
FOSB
|
FBJ murine osteosarcoma viral oncogene homolog B
|
|
chrX_+_70765479
|
1.497
|
|
OGT
|
O-linked N-acetylglucosamine (GlcNAc) transferase (UDP-N-acetylglucosamine:polypeptide-N-acetylglucosaminyl transferase)
|
|
chr10_-_77161136
|
1.496
|
|
ZNF503
|
zinc finger protein 503
|
|
chr2_+_171571936
|
1.487
|
|
|
|
|
chr21_-_33074101
|
1.478
|
|
SCAF4
|
SR-related CTD-associated factor 4
|
|
chr13_-_60325073
|
1.459
|
|
|
|
|
chr3_+_128532267
|
1.456
|
|
|
|
|
chr13_+_24144402
|
1.455
|
NM_001204459
NM_148957
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19
|
|
chr5_-_39394423
|
1.440
|
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr11_-_83984230
|
1.430
|
NM_001142700
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr3_+_63884074
|
1.408
|
NM_001177387
|
ATXN7
|
ataxin 7
|
|
chr4_+_74606222
|
1.404
|
NM_000584
|
IL8
|
interleukin 8
|
|
chr7_+_39017608
|
1.400
|
NM_001166018
NM_007252
|
POU6F2
|
POU class 6 homeobox 2
|
|
chr7_-_41742666
|
1.392
|
NM_002192
|
INHBA
|
inhibin, beta A
|
|
chr18_+_32073245
|
1.392
|
NM_001198945
NM_001392
NM_032975
NM_032979
|
DTNA
|
dystrobrevin, alpha
|
|
chr4_+_88571453
|
1.392
|
NM_001079911
NM_004407
|
DMP1
|
dentin matrix acidic phosphoprotein 1
|
|
chr10_-_77161424
|
1.374
|
NM_032772
|
ZNF503
|
zinc finger protein 503
|
|
chr1_+_46379258
|
1.373
|
|
MAST2
|
microtubule associated serine/threonine kinase 2
|
|
chr17_-_49124128
|
1.345
|
NM_001251971
|
SPAG9
|
sperm associated antigen 9
|
|
chr15_+_57511616
|
1.344
|
NM_207040
|
TCF12
|
transcription factor 12
|
|
chr3_+_157154579
|
1.341
|
NM_002852
|
PTX3
|
pentraxin 3, long
|
|
chr8_+_32579350
|
1.334
|
NM_001159996
|
NRG1
|
neuregulin 1
|
|
chr3_-_114102488
|
1.310
|
NM_001164342
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr17_-_8063946
|
1.308
|
|
VAMP2
|
vesicle-associated membrane protein 2 (synaptobrevin 2)
|
|
chr8_+_92261515
|
1.286
|
NM_052832
NM_134266
|
SLC26A7
|
solute carrier family 26, member 7
|
|
chr3_-_87325611
|
1.276
|
NM_000306
NM_001122757
|
POU1F1
|
POU class 1 homeobox 1
|
|
chr4_+_160188997
|
1.276
|
NM_014247
|
RAPGEF2
|
Rap guanine nucleotide exchange factor (GEF) 2
|
|
chr3_-_47933997
|
1.271
|
|
MAP4
|
microtubule-associated protein 4
|
|
chrM_+_10361
|
1.268
|
|
|
|
|
chr3_+_156394452
|
1.267
|
NM_001184718
|
TIPARP
|
TCDD-inducible poly(ADP-ribose) polymerase
|
|
chr9_+_2717525
|
1.256
|
NM_133497
|
KCNV2
|
potassium channel, subfamily V, member 2
|
|
chr5_-_124080773
|
1.255
|
NM_020747
|
ZNF608
|
zinc finger protein 608
|
|
chr2_-_211018294
|
1.247
|
|
C2orf67
|
chromosome 2 open reading frame 67
|
|
chr3_+_115342302
|
1.238
|
|
GAP43
|
growth associated protein 43
|
|
chr1_-_94586678
|
1.230
|
NM_000350
|
ABCA4
|
ATP-binding cassette, sub-family A (ABC1), member 4
|
|
chr1_-_100643770
|
1.226
|
NM_144620
|
LRRC39
|
leucine rich repeat containing 39
|
|
chr22_-_38506532
|
1.221
|
|
BAIAP2L2
|
BAI1-associated protein 2-like 2
|
|
chrM_+_1670
|
1.220
|
|
|
|
|
chr14_+_55168831
|
1.219
|
|
SAMD4A
|
sterile alpha motif domain containing 4A
|
|
chr12_+_50869531
|
1.217
|
|
LARP4
|
La ribonucleoprotein domain family, member 4
|
|
chr11_-_84634437
|
1.206
|
NM_001364
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr6_-_56716713
|
1.206
|
|
DST
|
dystonin
|
|
chr3_+_141043054
|
1.203
|
NM_001080412
|
ZBTB38
|
zinc finger and BTB domain containing 38
|
|
chr18_+_32073343
|
1.202
|
|
DTNA
|
dystrobrevin, alpha
|
|
chr1_+_151739130
|
1.192
|
NM_016178
|
OAZ3
|
ornithine decarboxylase antizyme 3
|
|
chr11_-_84634219
|
1.186
|
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr15_-_70390229
|
1.179
|
NM_001105192
NM_005078
NM_020908
|
TLE3
|
transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila)
|
|
chr3_+_151488214
|
1.170
|
|
LOC201651
|
arylacetamide deacetylase (esterase) pseudogene
|
|
chrM_+_2211
|
1.169
|
|
|
|
|
chr10_-_77161539
|
1.159
|
|
ZNF503
|
zinc finger protein 503
|
|
chr9_+_103340335
|
1.154
|
NM_001018116
|
MURC
|
muscle-related coiled-coil protein
|
|
chr3_+_25470067
|
1.146
|
|
RARB
|
retinoic acid receptor, beta
|
|
chrM_+_10704
|
1.135
|
|
|
|
|
chr10_-_75335415
|
1.132
|
NM_152586
|
USP54
|
ubiquitin specific peptidase 54
|
|
chr15_-_70390226
|
1.123
|
|
TLE3
|
transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila)
|
|
chr13_+_97928414
|
1.120
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr12_-_99548867
|
1.108
|
NM_001204067
|
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B
|
|
chr20_+_32150139
|
1.106
|
NM_001039709
NM_005093
|
CBFA2T2
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 2
|
|
chr4_-_74088773
|
1.104
|
|
ANKRD17
|
ankyrin repeat domain 17
|
|
chr11_+_124481451
|
1.097
|
NM_052959
|
PANX3
|
pannexin 3
|
|
chr7_-_27205075
|
1.086
|
|
HOXA9
|
homeobox A9
|
|
chr6_-_149806116
|
1.082
|
NM_207360
|
ZC3H12D
|
zinc finger CCCH-type containing 12D
|
|
chr4_+_74286846
|
1.070
|
|
ALB
|
albumin
|
|
chr2_+_33359607
|
1.066
|
NM_000627
NM_001166264
NM_001166265
NM_001166266
|
LTBP1
|
latent transforming growth factor beta binding protein 1
|
|
chr6_+_87964925
|
1.059
|
|
ZNF292
|
zinc finger protein 292
|
|
chr2_-_133539627
|
1.057
|
|
NCKAP5
|
NCK-associated protein 5
|
|
chr10_-_14372807
|
1.049
|
NM_018027
|
FRMD4A
|
FERM domain containing 4A
|
|
chr22_+_39101954
|
1.047
|
|
GTPBP1
|
GTP binding protein 1
|
|
chr4_-_153273683
|
1.047
|
NM_018315
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chr12_-_71031174
|
1.038
|
NM_001109754
|
PTPRB
|
protein tyrosine phosphatase, receptor type, B
|
|
chr7_-_16505292
|
1.038
|
NM_015464
|
SOSTDC1
|
sclerostin domain containing 1
|
|
chr13_-_41593491
|
1.033
|
NM_172373
|
ELF1
|
E74-like factor 1 (ets domain transcription factor)
|
|
chr17_-_63009302
|
1.033
|
|
GNA13
|
guanine nucleotide binding protein (G protein), alpha 13
|
|
chrM_+_11427
|
1.031
|
|
|
|
|
chr6_+_7579692
|
1.017
|
|
DSP
|
desmoplakin
|
|
chr6_-_138189313
|
1.013
|
|
|
|
|
chr7_-_155601757
|
1.011
|
|
SHH
|
sonic hedgehog
|
|
chr4_+_118955499
|
1.001
|
NM_004784
|
NDST3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3
|
|
chr7_-_44580857
|
1.000
|
NM_001101648
NM_013389
|
NPC1L1
|
NPC1 (Niemann-Pick disease, type C1, gene)-like 1
|
|
chr13_-_41556188
|
0.996
|
|
ELF1
|
E74-like factor 1 (ets domain transcription factor)
|
|
chr13_+_97874540
|
0.993
|
NM_144778
NM_207304
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr6_-_56507585
|
0.983
|
NM_001723
NM_015548
|
DST
|
dystonin
|
|
chr7_-_37025696
|
0.982
|
|
ELMO1
|
engulfment and cell motility 1
|
|
chr10_+_63808968
|
0.981
|
NM_001244638
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like)
|
|
chr12_-_117319231
|
0.980
|
NM_003806
|
HRK
|
harakiri, BCL2 interacting protein (contains only BH3 domain)
|
|
chr14_-_81864726
|
0.970
|
NM_033104
|
STON2
|
stonin 2
|
|
chr1_+_167189948
|
0.967
|
NM_001198786
NM_002697
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr20_-_55934877
|
0.964
|
NM_001190472
|
MTRNR2L3
|
MT-RNR2-like 3
|
|
chr8_+_77593522
|
0.962
|
|
ZFHX4
|
zinc finger homeobox 4
|
|
chr19_+_13135368
|
0.957
|
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr3_-_105421044
|
0.953
|
|
CBLB
|
Cas-Br-M (murine) ecotropic retroviral transforming sequence b
|
|
chr6_-_41703938
|
0.953
|
NM_001167827
|
TFEB
|
transcription factor EB
|
|
chr13_-_41593405
|
0.951
|
|
ELF1
|
E74-like factor 1 (ets domain transcription factor)
|
|
chr11_-_83393436
|
0.941
|
NM_001142702
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr11_-_85338313
|
0.941
|
NM_001142699
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr13_+_78315294
|
0.940
|
NM_001242869
NM_001242870
NM_001242871
NM_144595
|
SLAIN1
|
SLAIN motif family, member 1
|
|
chrX_+_69488154
|
0.938
|
NM_004312
|
ARR3
|
arrestin 3, retinal (X-arrestin)
|
|
chr4_-_82393025
|
0.934
|
NM_152545
|
RASGEF1B
|
RasGEF domain family, member 1B
|
|
chrX_-_76814217
|
0.929
|
|
ATRX
|
alpha thalassemia/mental retardation syndrome X-linked
|
|
chr7_+_116395440
|
0.926
|
|
|
|
|
chr12_-_26277807
|
0.925
|
NM_030762
|
BHLHE41
|
basic helix-loop-helix family, member e41
|
|
chr10_+_63661012
|
0.923
|
NM_032199
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like)
|
|
chr5_-_39424930
|
0.922
|
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr17_-_46651809
|
0.919
|
NM_002146
|
HOXB3
|
homeobox B3
|
|
chr9_-_110251308
|
0.919
|
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr4_-_82392971
|
0.917
|
|
RASGEF1B
|
RasGEF domain family, member 1B
|
|
chr10_-_62149487
|
0.917
|
NM_020987
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr2_-_198540583
|
0.913
|
NM_144629
|
RFTN2
|
raftlin family member 2
|
|
chr12_-_102874373
|
0.912
|
NM_000618
NM_001111283
NM_001111285
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr14_+_75745480
|
0.910
|
NM_005252
|
FOS
|
FBJ murine osteosarcoma viral oncogene homolog
|
|
chr1_-_213020947
|
0.909
|
NM_001024601
|
C1orf227
|
chromosome 1 open reading frame 227
|
|
chr8_+_77593506
|
0.906
|
NM_024721
|
ZFHX4
|
zinc finger homeobox 4
|
|
chrM_+_14601
|
0.903
|
|
|
|
|
chr10_-_126716452
|
0.899
|
NM_022802
|
CTBP2
|
C-terminal binding protein 2
|
|
chr7_+_102553343
|
0.896
|
NM_001031692
NM_005824
|
LRRC17
|
leucine rich repeat containing 17
|
|
chr6_-_56716412
|
0.895
|
|
DST
|
dystonin
|
|
chr4_+_88529680
|
0.895
|
NM_014208
|
DSPP
|
dentin sialophosphoprotein
|
|
chr1_+_78470593
|
0.894
|
NM_007034
|
DNAJB4
|
DnaJ (Hsp40) homolog, subfamily B, member 4
|
|
chr12_-_51611476
|
0.894
|
|
POU6F1
|
POU class 6 homeobox 1
|
|
chr18_+_3447607
|
0.890
|
NM_173207
|
TGIF1
|
TGFB-induced factor homeobox 1
|
|
chr16_+_69984809
|
0.890
|
NM_182619
|
CLEC18C
CLEC18A
|
C-type lectin domain family 18, member C
C-type lectin domain family 18, member A
|
|
chr3_-_25683825
|
0.886
|
|
TOP2B
|
topoisomerase (DNA) II beta 180kDa
|
|
chr6_-_53530505
|
0.880
|
NM_001003760
|
KLHL31
|
kelch-like 31 (Drosophila)
|
|
chr8_-_87755880
|
0.868
|
NM_019098
|
CNGB3
|
cyclic nucleotide gated channel beta 3
|
|
chr7_-_87342569
|
0.866
|
NM_000927
|
ABCB1
|
ATP-binding cassette, sub-family B (MDR/TAP), member 1
|
|
chr2_+_54785453
|
0.865
|
NM_178313
|
SPTBN1
|
spectrin, beta, non-erythrocytic 1
|